This is the second article in a series called Enzymes for Brewers. The first article covered catalysts and protein structure. This article — on enzyme function — is mostly background material, needed to understand the more brewing-relevant information to come.
In this post, I’ll explain the basics of how proteins work. But first, since much of this may be new to some readers, let’s quickly review the information in the first installment of this series.
A catalyst is a molecule that speeds the rate of a chemical reaction, but is not used up in that reaction. Enzymes are catalysts that are biological molecules. Most enzymes are proteins, but a few RNA enzymes exist. Proteins are long strands of amino acids, joined end to end by peptide bonds. These molecules are “built” on a cellular organelle called a ribosome.
[Brief tangent: In biochemistry lingo, proteins are translated from the code of a messenger RNA (mRNA) molecule into the specific sequence of amino acids that comprise the protein. The mRNA (which itself is a sequence of molecules called nucleotides) was transcribed from a gene — a strand of DNA (which is also comprised of a string of nucleotides). In short, DNA sequences are transcribed into RNA sequences, which are in turn translated into protein sequences. Here’s a website that gives an interactive example. Also, see Wikipedia’s pages on transcription and translation for how the information from DNA (a sequence of nucleotides, in a helical molecule) gets turned into a protein (a sequence of amino acids, often formed into a globular molecule).]
Protein strands are produced starting at the N-terminus, the end of the strand that terminates with an amine group (–NH2) on the first amino acid, and proceeds towards the C-terminus. The C-terminus is the end of the protein and a carboxyl group (–COOH) is hanging off the last amino acid added to the protein strand. Because the N-terminal end of the protein starts to fold while the protein starnd is still be added to, and because many proteins fold with the help of other proteins (called chaperones), denatured proteins frequently cannot refold into their active form. [To make things even more complicated, some proteins are modified (by other enzymes) after they originally fold. That’s another reason that, once the 3-D structure of a protein is disrupted, in many cases it cannot be regained.]
Protein Functions
Not all proteins are enzymes. There are structural proteins, cell signaling proteins, and carrier proteins as well as metabolic enzymes. Structural proteins, like collagen, are frequently fibrous. Cell signaling proteins are usually found on the surface of the cell. Carrier proteins, like hemoglobin, which carries iron in the blood of most vertebrates, transport molecules within a cell or organism.
Enzymes are Specific and Effective
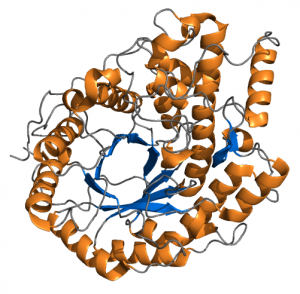
Most proteins that act as enzymes are globular. In yeast, the average length of a protein is 466 amino acids, but enzymes can vary from around 60 to around 25,000 amino acid residues. In solution, the 3-D structure of an enzyme is deformable, as the overall shape is mostly held together by hydrogen bonds, a fairly weak association between atoms in adjoining strands. The shape may additionally be stabilized by disulfide bonds, formed when two sulfur-containing amino acids in the protein strand are brought close together in the folded state of the protein.
Enzymes are very specific, most often catalyzing only one chemical reaction within the cell. A few catalyze more than one reaction (among similar substrates), or can catalyze different reactions in different environments. Enzymes frequently increase the rate at which a chemical reaction occurs by over a million fold. (And that’s the actual comparison of measured rates.)
So how does an enzyme — essentially a “squishy” ball of a molecule — catalyze chemical reactions?
Shape Determines Function
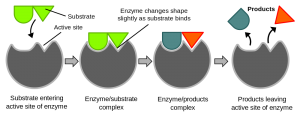
The function of an enzyme is determined by its shape. The chemical reaction an enzyme catalyzes occurs in its “active site.” In most proteins, the active site is a pocket or cleft in the molecule. The active site frequently occupies only a small fraction of the enzyme’s overall volume. Enzymes work by binding the substrate at the active site and causing the reaction to occur.
The oldest model of enzyme function is the lock and key model, proposed by Emil Fisher in 1894. He proposed that the shape of the active site of an enzyme was complementary to the shape of the substrate. In other words, the substrate was like a puzzle piece with a tab extending from it and the enzyme was a puzzle piece with the appropriate pocket for the tab to fit into. If the enzyme catalyzed a reaction of two molecules — which we’ll call A and B into a product, C — the shapes of A and B would lead them to fit into the enzyme. If the active sites for the A and B molecules were right next to each other, A and B would be brought into close proximity, and in the correct orientation, for the reaction to occur. This was a remarkable theory given that nothing was known about protein structure at the time, and still serves as a teaching model to introduce students to enzyme function. However, it has been improved upon.
In 1958, Daniel Koshland proposed a modification of this idea. In Koshland’s model, the substrate or substrates don’t initially fit into the active site. However, after their initial contact, the enzyme and the substrate interact so that they eventually fit together forming an enzyme-substrate complex. In this structure, the substrate is mostly surrounded by the enzyme’s active site. (Remember that enzymes are like squishy balls and their substrate(s) may be easily deformed by contact.) This is called the induced fit model.
Catalyzing the Reaction
So, if you imagine the enzyme as a wiggly blob that ends up “grasping” or partially enclosing the substrate or substrates, how does the enzyme catalyze the reaction? There are several ways this can occur. For starters, an enzyme can bring two substrates together in the correct orientation for the reaction between them to occur. This alone doesn’t mean that it will occur, however, as there may be an activation energy that has to be overcome. And also, this doesn’t explain how reactions that split a substrate into two molecules is accomplished.
A reaction can be catalyzed in several ways. In the case where an enzyme splits a substrate, an enzyme can flex or twist a substrate molecule, such that the activation energy for the reaction to occur is lowered. The local electrical charge in the region of the active site may make the reaction more likely to occur by providing an electron donor or an electron acceptor. An enzyme can catalyze a reaction by providing an energetically favorite intermediate for the reaction. For example, when starch is synthesized in plants (including barley), the joining of one glucose molecule to another occurs via an intermediate reaction between glucose (with a phosphate molecule attached) and ATP, forming an ADP-glucose molecule (and a pyrophosphate molecule). This molecule then reacts with the glucose, adding one glucose molecule to the starch chain and releasing ADP.
So, the enzyme brings the substrates together and either deforms them, supplies them with electron donors or receptors, provides them a stable intermediate pathway for the reaction, or any combination of these such that the reaction occurs.
Summary and Brewing Example
So, you can think of an enzyme as a squishy, wiggly ball made from a string of amino acids. It floats around in the cytoplasm of the cell and, if by chance it bumps into its substrate, it ends up partially engulfing it in its active site. Once the enzyme-substrate complex is formed, the relevant molecules are held in the right position, in the right environment, for the reaction to take place.
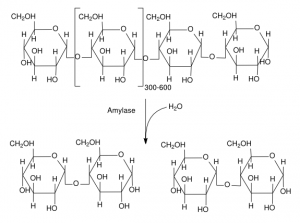
In brewing, one of the most important enzyme-catalyzed reaction is the reduction of starch to simpler sugars. One enzyme that participates in this is beta-amylase. This enzyme, once folded and processed, is a string of 498 amino acids. In the mash, it floats free in the wort. Since enzymes work solely as a function of their shape, they can work in any aqueous solution (presuming the salt content, pH, etc., is in the right range). When beta-amylase bumps into the reducing end of an amylose (starch) molecule, it “gloms onto” the starch. Once the enzyme-starch complex is formed, the enzyme (somehow) arranges it so a water molecule can be positioned to cause the hydrolysis of a bond between two glucose residues in the starch.
After the reaction occurs, the enzyme releases the remaining section of the starch strand and a maltose molecule. The atoms of the water molecule are now apart of the starch strand and maltose molecule. The beta amylase enzyme again floats free, the laws of physics and chemistry returning it to original confirmation and — if it happens to bump into another starch molecule — it can catalyze the reaction again.
Fun Fact
Did you know that you can get over 100% extract efficiency? Extract efficiency is calculated by taking the dry weight of the starch in your grain and comparing it to the dry weight of the resulting sugars (inferred from the specific gravity reading). The weight of the sugars includes both the weight of the starch plus two hydrogens and one oxygen from every water molecule. If you were to completely degrade the starch in your grain, the resulting sugars would weigh more than the starch the came from. The water weight would push your efficiency above 100%. In breweries, this level of extract efficiency is never achieved. However, it has been achieved in lab mashes.
In upcoming articles, will examine how enzymes are regulated and — in the most brewing-relevant article — the kinetics of enzymes.
—
Related articles
Tannins for Brewers (The Basics)

Speak Your Mind