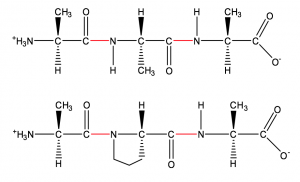
Figure 1: Simplified structures of two peptides. The top peptide is made from three alanines, and on the bottom is the peptide alanine-proline-alanine. The peptide bonds are shown in red.
This is the first part in a 3-part series, written by Chris Hamilton of Hillsdale College. The second and third parts will be posted tomorrow (Thursday) and the next day (Friday). Hear an interview and tasting of samples on Basic Brewing Radio – September 4, 2014.
Gluten is a popular topic lately when it comes to food and beer, both due to an increased awareness of Celiac disease and popular diets which eschew any gluten containing food or beverages. Chris Colby wrote the article, Gluten and Brewing, back in November of 2013 that summarizes many of the challenges in brewing a gluten-free beer. The part of gluten and hordein (the barley equivalent of gluten) that can cause problems for some people is the protein gliadin.
As a biochemist who studies enzymes, one of my main areas of research the past seven years has been with enzymes called proteases that breakdown proline-rich proteins. Proline is unique amongst amino acids found in proteins; rather than forming the typical peptide bond it forms a ring structure with its amino nitrogen (see Figure 1). Essentially, this means it is a tough bond to break, and the proteases found in grain and in the human digestive system are unable to efficiently break the peptide bonds on both sides of a proline. Gliadin proteins are broken down during digestion into several different peptides containing proline, these peptides are what triggers the immune response in people with celiac disease.
A very specific class of enzymes called prolyl endopeptidases are able to catalyze the breakdown of proteins after proline residues. It was only in 2002 that several researchers proposed the possibility of using these enzymes from bacteria and fungi to break down proteins like gliadin into very small peptides and/or free amino acids. 1,2 A number of my students have characterized prolyl endopeptidases from various species with hopes of finding a good candidate for further development.3,4
The most recent recommendation from the Codex Alimentarious Commission has been that products with less than 20 parts per million (ppm) of gluten can be considered gluten free. The U.S. Food and Drug Administration has adopted nearly the same policy for labeling of gluten free foods, so as of August 5, 2014 any food product made from a gluten containing grain (wheat, barley, rye, or their hybrids) that has been treated to remove gluten may be labeled as gluten free if it contains less than 20 ppm of gluten.
Clarity Ferm
White Labs sells a product known as Clarity Ferm, which is made from a product called Brewer’s Clarex (manufactured by DSM). Brewer’s Clarex is a solution that contains a purified prolyl endopeptidase from Aspergillus niger — aka black mold. But don’t worry there’s no mold in the solution, just purified proteins. DSM and then White Labs initially marketed their products to reduce chill haze in beer (hence the “Clarity” and “Clarex” in the product name). Proteins bind to polyphenols in beer to cause haze at low temperatures, if you add an enzyme that breaks down these proteins then you can eliminate or reduce the chill haze. Since this enzyme is a prolyl endopeptidase, a side-effect of using Brewer’s Clarex and Clarity Ferm is that the gliadin and hordein (gluten) are broken down as well.
Charlie Papazian did an experiment in 2009 with Brewers Clarex that piqued the interest of many homebrewers. The finished beer was tested for gluten and was found to have less than 5 ppm of gluten, however the ability to detect fragments of gluten protein back then was not readily available. So there was uncertainty in the findings.
It’s only in the last few years that White Labs has marketed Clarity Ferm as being able to reduce gluten in beer. So there is some real evidence out there that the enzyme in Clarity Ferm will break down gliadin and similar proteins, BUT is it enough? Does is break down all the gliadin? Is it broken down completely or are there large potentially toxic protein fragments still around? The product is now sold in 10 mL vials, which they recommend is suitable to reduce the gluten in a 5-10 gallon batch of beer.
Testing for Gluten
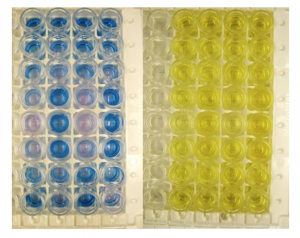
Figure 2: ELISA plates. Left plate shows color after adding the samples and the substrate, the right plate is shown after stopping the reaction. The absorbance of light at 450 nm is used to measure the amount of product formed, which allows for the quantification/detection of gluten and its fragments.
Fortunately, a test for peptides from gliadin and similar proteins is now available and it is specifically designed to test food and beverages. The RIDASCREEN® Gliadin competitive kit from R-Biopharm is an enzyme-linked immunosorbent assay (or ELISA). This means it uses anti-bodies that are highly sensitive at detecting small fragments of gliadin. Samples are loaded into small wells on a plate (like mini test tubes) and mixed with either standards or the sample being tested (in our case beer). The nice thing about this test is that it was specifically designed with the idea of testing food and beverages including beer. In the assay, if any gliadin peptides bind to the antibodies they will cause a specific color change on the plate. This change can be measured you a special instrument called a UV-Vis spectrophotometer. To quantify the amount of gliadin in the beer, it is compared to standards which are run at the same time on the plate in duplicate (Figure 2).
References
3. Banach, S. (2009) Senior Thesis. Purifying novel prolyl oligopeptidases to observe proteolysis including gluten degradation for Celiac Disease. (Research Advisor: Christopher Hamilton) Hillsdale College, Hillsdale, MI.
4. Keyes, S. (2011) Senior Thesis. The cloning and initial characterization of a prolyl endopeptidase of Spriosoma linguale. (Research Advisor: Christopher Hamilton) Hillsdale College, Hillsdale, MI.
—
Related articles

Speak Your Mind